Microsatellite instability (MSI) is listed in genetic test reports, but it’s a name guaranteed to baffle, and — unlike the nearly all the other entries — it’s not a mutated gene.
We talk here about what microsatellite instability is, what it indicates, and why it wears that name.
Microsatellites are also the basis of paternity tests and DNA identification in law enforcement. Naturally we’ll talk about that too.
First, the name
The genetic code was famously “cracked” in 1966, but there’s plenty of puzzlement left. The code told us how DNA specifies proteins, but less than 2% of DNA makes protein. That leaves stretches where, like Stonehenge, it’s not obvious what was intended.1
Within this mystery world are back-to-back repeated sequences called tandem repeats.2 The repeating motif can vary in size from thousands of nucleotides3 to a long stretch of a single nucleotide.
Satellite is another name for such a repeating sequence. It comes from test tubes whose contents looked like this:
Above the main section of DNA (labeled “Main genomic fraction”) appear “satellite” bands.
What does that have to do with repeats?
Each of the four DNA nucleotides weighs a different amount, and an adenine-thymine pair weighs less than a guanine-cytosine pair. If you chop DNA coarsely (this was the 1960’s), pieces that contain a sizeable repeating sequence are likely to have a weight different from the average, and if you whirl the tube in a centrifuge, the lighter sequences will appear higher. You can see the highest band is predominantly A and T.
As techniques improved, researchers could distinguish shorter repeating motifs that they named minisatellites, and ones still shorter got named microsatellites — or short tandem repeats. The repeating block of a microsatellite has one to roughly six nucleotides (there’s no standard maximum).
Next, the instability
Let’s tie this in with prostate cancer.
It turns out that the simplest tandem repeats — the ones consisting of one repeated base pair — are the most useful in cancer diagnosis. It’s easiest for cellular machinery to lose its place in a string like that.
Like genes, repeats occur at fixed locations in specific chromosomes. (They also, like genes, have names.) But the length of the repeat can vary from person to person. BAT26 is always a repetition of A’s (BAT stands for Big Adenine Tract), but my BAT26 might have more or fewer A’s than yours.4
But in every one of our cells the BAT26 length will be the same.
Unless one of us has microsatellite instability. Then his tumors will have more or fewer A’s than his normal cells do.
A diagnostic MSI test will examine at least five microsatellite sites. MSI is deemed high — MSI-H — when instability is reported in more than two.
MSI-L — instability in a single location — is the Gleason 6 of MSI, with papers titled Does MSI-low exist? Standard practice is to ignore it, so MSI means MSI-H.
The absence of MSI is microsatellite stable, MSS.
MSI is associated with colorectal cancer more often than prostate cancer —about 3%–5% of advanced prostate cancer patients have MSI versus about 15% of colon cancer patients. As a result, guidelines that specify which microsatellites to test (famously, the Bethesda panel) are written with colorectal cancer in mind and aren’t ideal for prostate patients; you may want to ask your doctor when discussing testing.
How MSI is tested
The search for MSI classically begins with PCR — the same PCR we met in COVID-19 testing. PCR (polymerase chain reaction) makes many copies of a small stretch of DNA, harnessing the biological machinery that cells already use for copying DNA.
Further processing is done to separate the microsatellites by length5, yielding results like these:
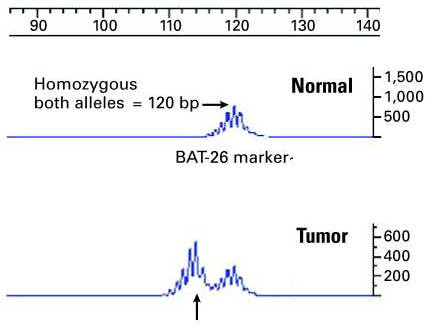
The display for the tumor includes a shifted peak6, indicating a different microsatellite length — in other words, microsatellite instability.
The genes behind MSI: mismatch repair
We mentioned that MSI itself isn’t a mutation.
It’s indirect evidence of a loss of at least one of four genes tasked with DNA repair — MLH1, MSH2, MSH6, and PMS2 7.
Cancer is the product of mutation, and broken repairs mean mutations can wildly proliferate. Changes in microsatellite length — particularly in mononucleotide satellites — are part of this chaos, and they are easy to spot.
Finding dMMR
The gene loss can also be found directly.
The four genes are known collectively as mismatch repair genes; the defect is called dMMR, for defective mismatch repair.
You don’t need sequencing to find which genes are defective — you can see it in a microscope.
The test uses stains that change color in the presence of a specified protein. The stains in the slides above detect the proteins expressed by each of the four genes. Brown means the protein is present. In this patient, the genes MLH1 and PMS2 are OK, MSH2 and MSH6 are not.
The very subtle brown in the MSH2 and MSH6 sides is a check that the test is working. An unrelated part of the cell is also stained; a slide without any brown means something is wrong with the test, not that there’s a genetic loss.
Testing of this kind has been done since the 1940’s. It goes by the impressive name immunohistochemistry8 (IHC).
Pathologists use it ubiquitously. My lung biopsy tissue, for instance, was stained for NKX3.1 and PSA (which were positive, indicating it was prostate cancer) and TTF-1 (negative, indicating it wasn’t lung cancer).
Yet another clue: mutational burden
If repair loss has allowed mutations to run amok, DNA sequencing can directly count them. High tumor mutational burden (TMB-H) signals a likelihood of defective mismatch repair. “High” is defined differently by different researchers, but a sample definition is 17 mutations per megabase — a million base pairs — after examining at least 1.2 million. Ten mutations per megabase is the FDA’s threshold for prescribing pembrolizumab.
The DNA locations examined for tumor mutational burden don’t overlap the set of examined microsatellites. TMB counts protein-altering modifications, but the MSI microsatellites that are tested are from parts of the DNA that do not create proteins.
Most often clinicians will sequence a subset (a panel) of genes rather than sequencing every protein-coding gene in the tumor. The FoundationOne CDx gene panel has explicit FDA approval as a biomarker for prembrolizumab.
Three equivalent markers?
The FDA allows prescribing pembrolizumab in the presence of any of the three indications — MSI-H, dMMR, or TMB-H. Are they equivalent?
Dr. Emmanuel Antonarakis of the University of Minnesota has said that success with pembrolizumab is likely only for patients showing all three.
He discusses it in a December 2021 Urology Today video (I haven’t seen it elsewhere):
So, when you see all three, a loss of function — mismatch repair mutation — and a high TMB, and the microsatellite instability, you begin to believe that this is a true pembrolizumab-sensitive tumor. Oftentimes you get tricked and you see one of the three or two of the three, and then you are kind of stuck because you sort of want to give pembrolizumab, but you don't know if that patient is going to respond.
Isn’t BRCA2 a DNA repair mutation?
Several pathways in addition to mismatch repair are devoted to repairing DNA (the umbrella term is DDR, for defective-DNA repair). Cells spend as much effort checking and maintaining DNA as they do copying it.
The BRCA2 biomarker arises in a different repair pathway, homologous recombination repair (HRR), and there are still others.9
So why immunotherapy?
Without immune checkpoint inhibitors — the class of drug that pembrolizumab belongs to — we wouldn’t much care about any of these biomarkers. They’re immunotherapy drugs. How can an immunotherapy drug be useful for DNA mismatch repair?
The short explanation centers on the high rate of mutation, which produces many antigens that elicit a large-scale immune response. Cancer protects itself with the same safety switch that normal cells use to prevent autoimmune attack, and drugs like pembrolizumab defeat the switch so the attack can go forward.
Pembro’s applicability in MMI-H/dMMR/TMB-H wasn’t discovered until the drug had already been approved for other cancers. Researchers turned an apparent failure in a colorectal cancer trial (success in only one of 33 patients) into a breakthrough by recognizing that the patient had mismatch-repair deficiency.
It is the first drug to have received tissue/site-agnostic approval from the FDA — under certain conditions, it can be prescribed for any cancer positive for MSI-H or dMMR.
Much more can be said about pembrolizumab. A closer look at it and other immunotherapies awaits a future post.
MSI → CSI
And now the bonus discussion on DNA fingerprinting.
Recall that people may have different repeat counts in a given microsatellite, but every person will have that same count in all cells. (We’ve left microsatellite instability and are now talking about microsatellites generally.)
DNA fingerprinting uses a suite of microsatellites selected to have lengths that vary greatly among individuals. The number of microsatellites and their variability produce so many combinations that no two people are likely to match in every microsatellite.
In fact, no two people are likely to match even half the satellites, which is where maternity/paternity testing comes in. Microsatellite lengths are inherited. Roughly half a person’s counts come from each parent, so a match of roughly 50 percent with a parent’s DNA can establish parenthood.
Stonehenge is 4,500 years old and DNA is 4 billion years old. Stonehenge can wait.
Though tandem suggests a nucleotide conspiracy, in this context it means only that repetitions appear back to back.
Nucleotide: Recall that DNA is made up of sequences of just four ingredients (nucleotides) whose names are abbreviated A, C, G, and T (for adenine, cytosine, guanine, and thymine). Recall, too, that each DNA rung has a pair of these — A always with T, or G always with C. The two together are a base pair, abbreviated bp.
In at least 99% of ethnic Europeans BAT26 has 26 adenines, but different lengths (eg, 15, 20, 22, 23) are seen in up to 25% of ethnic Africans, including African Americans. This was discovered in 1999 and shows the importance of research on ethnically diverse populations.
Via a technique known as capillary electrophoresis. In somewhat the same way the Covid test is called a PCR test when it clearly is more than PCR, the MSI + capillary electrophoresis test is often just called a PCR test as well.
The noisy mini-peaks aren’t in the tissue — they’re an artifact (stuttering) of PCR, which is having the same kind of trouble replicating a single-nucleotide string that cells do.
Genes in medical literature are named in italic. Each gene makes a protein, and the protein frequently has the same name as the gene, but isn’t italicized. Seeing an unfamiliar abbreviation in italics is a hint that it’s a gene.
Immuno because it uses manufactured antibodies that bind to a particular protein; histo from histology, or cellular anatomy.
Thus I’d imagine tumor burden could also reflect failures in a non-MMR pathway, but I don’t know.